Interstellar Contest on Fuun Programming 2007
Postmortem
On July 20–23, 2007 we participated at the ICFP Programming Contest. Our group consisted of six people, four from the chair of Programming Language and two from the group of Softwaretechnik at the University of Freiburg. For me it was the first contest of this kind with such a strict deadline of 72 hours. It was a spontaneous decision; Martin asked 24 hours before the contest started if someone wanted to join. But you cannot prepare for such a contest anyway :)
Day 1
Friday at noon I downloaded the contest description. The contest task was to repair the genetic material of an alien called Endo. He has a special genetic structure consisting of the bases I,C,F, and P, which encodes a programming language based on regular expressions. The output of this process, the RNA, consisted of drawing primitives, that allowed the generation of an image displaying Endo's shape. The goal was to change Endo's genes by adding a prefix in front of it, that should modify the process to produce a different image where Endo lives happily in the shape of a cow.
I read the first part of it, the part describing DNA to RNA conversion before we met to decide how to tackle this. We split into small groups of two people each. Since we favoured different programming languages I decided to program a DNA to RNA conversion in C, while the others splitted in two groups, one programming the DNA to RNA conversion in ocaml and one the RNA to image conversion.
After we would have finished the programs we wanted to discuss the further route. One idea was to go backwards; to first translate the image into RNA and then the RNA in DNA. I suspected it was important to understand the DNA. In hindsight it turned out, my suggestion matched the expectations of the organizers, but the other "brute-force" approach would also have worked, as Jed Davis proved with his third place.
But we are not there, yet. After five hours I had the first C program running. I needed another half hour to fix the bugs. Then the dna2rna process ran, ... and ran ... and ran. So after ten minutes I checked in the first 9429 bytes of RNA. The rna to image group was long finished at the time and was eagerly awaiting something to test with. After 23 minutes, we had 29 kb of RNA. But there was almost nothing visible in the produced image. One could only see a bit of a sine curve that could belong to the sky background. ‘I already have 50,000 iterations, how many iterations is it going to take?’ ‘There was something in the last chapter of the documentation.’ Hmm, I did not read the second part of the manual carefully, so I looked it up: 1.8 Million iterations! My program was far too slow.
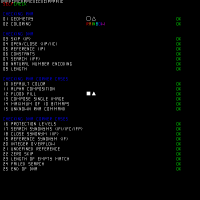
I then also tried the test prefix from the manual, which produced the complete result much faster. To our astonishment it produced a self-check. It also revealed a bug which could be easily fixed. The right hand side shows the picture after the bugs in the dna converter were fixed. The next few hours I spend optimizing the code and got it into the order of 250 iter/s, which means it could produce the source image in two hours. Certainly, this still was much too slow. But at least we had the complete source image RNA now. This was Friday evening at 10 PM.

So at the end of the first day after about twelve hours, we had the source image RNA, however, there was still a serious bug as can be seen in the image. I suspected it would be in the rna to image conversion. I spent some time at home to learn a bit of ocaml and actually could locate that bug at 1 AM. Now we had a perfect reproduction of the night image.
Day 2
I was too excited to sleep more than a few hours, so I planned the next
version of my dna processor. The contest description explicitily mentioned
that one has to make skips and appending unquoted patterns super-linear. In
my implementation skip was constant, but for appending I used memcpy. This
was also were almost all time was spent. So I thought of a way to fix this,
preferrably, without changing the underlying data structure, since all code
assumed the DNA was a plain char*. After inspecting the kind of
patterns I came to the conclusion that it may suffice to change the pattern
copy strategy: If the new destination of the pattern matched the old, no copy
is necessary. In some patterns a chunk of 7MB dna was quoted. For those one
could align the destination DNA so that only the remaining part of the DNA
needs to be copied. The pattern matching got quite complicated this way, but
it improved run-time. However, still most time was spent in memcpy. Also the
implementations was very buggy due to its complexity. At Saturday afternoon I
had buggy DNA converter of which I knew it cannot be made much faster without
a complete rewrite.
This meant I wasted more than 24 hours. The only good thing was that we had the RNA to analyse and that I had spent some time understanding the DNA conversion process and already found a few programming constructs like loops. Also having the green zone at a fixed place helped me enormously in interpreting the log files of the DNA conversion.
I did not mention, what the rest of our group did. Phillip and Markus were working on the ocaml version of the DNA processor. First they had the usual problems getting started, e.g. that ocaml could not handle large arrays (until our ocaml Guru told them how to use large arrays). After my debacle with the C program they started to work on a new data structure for the DNA using a tree. One of them programmed through until saturday morning programming a tree data structure for dna. However it did not run properly.
Annette and Stefan worked on the RNA to image conversion and had nothing to do until Friday evening, when the first rna was produced. Then Annette was working on analysing the RNA. She annotated most of it, determinining which object it draws.
After the whole debacle with my C program I decided to help he ocaml people with the tree based DNA converter. The code was already finished, so I helped bug tracing. I compared the output with the one produced by the C tool and we quickly could fix the remaining bugs. Now we finally had a converter that run in few minutes.
All the time I already read the logs of my C programs to understand the DNA process. I discovered the repeated pattern that computes the color for the night sky. It was produced by a subroutine that was called with certain numerical parameters as I could see from the log file. The numbers matched the number of produced RNA commands. So I started to write my first prefix, which changed these numbers:
IIPIPCCCCICCCCICIICCCCIIIICCPIICIPICICCCCCPIICIPPPFFFFFFFFFFFFFFFFFFFFFFFC FFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFC FFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFC FFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFCFFFFFFFFFFFFFFFFFFFFFFFFCFIIC
This changed the color of the sky as I had hoped. Of course, not a single pixel was correct and we still had to find out the right sky color, but it was a start. Actually I wrote a perl script to produce such prefixes. The perl script would do the task of converting human-readable patterns into Fuun DNA.
Now I started to automatise the task that Annette did manually: Search for addbmp, clip and compose RNA and split the RNA into small chunks, processing each separately to find out what each layer does. Feed everything into the image tool and look at the result. Writing the tool took some time, but running it took also about an hour, but the result was quite useful. This way I localized the darkening bitmap. I added a skip instruction that should jump over the RNA producing the opaque part of the color. Unfortunately, to skip the correct numbers of bytes one has to know how long the skip pattern is I used the following crude hack:
$pattern = pattern("!440").template("");
$len = length($pattern);
$pattern = pattern("!".(440-$len)).template("");
die "$len / $pattern" if ($len != length ($pattern));
print pattern("(!6642614)!$len").template("m0,0".$pattern);

On Sunday at 2 o'clock in the morning the darkness should vanish :). I could not wait submitting the prefix to the server. The result came quickly (the server was much faster than our ocaml tool). The prefix got a score of 3583460, 0.0000 % survival chance. Okay, there were some bugs in the prefix; it took some time too fix, especially since our tool was still a bit slow, and we did not want to upset the people in Utrecht by misusing their server for debugging our prefixes :). But after 50 minutes it worked in our tool. It fixed more than a third of the pixels in the image. I was so proud! We had no chance for the lightning contest, but maybe we are back now! At 2:53 AM we had our second prefix (left-hand image): Risk 2007350, survival chance 0.0002%. It was getting a bit frustrating.

Another big part of the image is the cargo box, so I fixed this. It turned out that it sufficed to change one RNA from magenta to orange. However, many teams already had a survival chance of 1.2719% with a prefix length of only 28, while our first patch was already 70 bases long. Was there a crucial hint we missed? Wait a minute, wasn't there a prefix in the self-test picture? In the mean time it disappeared because we fixed the bug in the RNA conversion, but luckily we have used a version control system. We quickly discovered the daylight switch prefix, combined it with our cargo-box prefix and at Sunday 4:43 AM we entered the top twenty with 2.9% survival chance. (I think at that time there was no score published for the top twenty teams, later the top area was shrinked to fifteen.) We never left this top area again.
IIPIFFCPICPCIICICIICIPPPPIICIIPIPIIICCCCIICCIICIIICIIICPIICPFIICIPPPFPIIC
Of course, we also found the manual pages at that moment. Actually we didn't get the hint with the index page immediately, but we knew how to manipulate numbers. So we just used brute-force to generate the manual pages from 1 to 100. At 5:40 AM I finally got the hint and the index page and the remaining manual pages listed there. At six o'clock we checked in a README for the other part of our group who went home early and I tried to get some sleep.
Day 3
I didn't sleep much and read the manual pages instead. I liked page 1729 as it revealed that all my previous analysis on the DNA process was correct. I knew at that time that the blue area was the stack, the green area the program and the red area the current function. I decided to write a program that gives out the call tree from the debug output of our DNA process. It was nice to see that there were exactly three calls to 3c870e:00372b, which the gene table denoted as appletree.
Page 8 inspired me to write a program to look out for structures like strings and polygons. I wrote a program to extract the polygons from DNA and Annette wrote a program to visualize them. I also extracted the texts and after doing some substitution cipher crack it was readable (I only found out after the contest that it was EBCDIC, however, the charset manual page was very helpful to quickly find the alphabet). The symbol table was very useful, however, I needed some time to guess how to get to the other pages. I found the symbol table also in the string dump but without the address and lengths. Much later I realized that the offset and length is in the dna at the same place.
So on sunday evening, we finally had a full symbol table, we had a call tree and we could start doing some serious DNA hacks. The first was to replace the apple tree function by some code that jumps to pear tree. After fixing an offset in our prefix perl script (didn't the old value work before?) it worked. So we submitted a new prefix, but the reported result was bad. We continued improving, but the contest system would give as ridiculously low points. Only after a while I noticed that someone of our team had checked in a new version of endo.dna with a fixed prefix into our repository. After reverting the change to endo.dna and putting the old offset into our perl script everything was fine. First we mainly removed objects that do not belong to the target image. We replaced the string ‘Morph Endo!’ but the space was only large enough for ‘Endo has mo’. This way it could also stand for Endo has moved away, since at that time there was no Endo in our picture :)
Later we started to call the functions which by their name would draw the
objects of the target image. It turned out that many of them would not work,
or do something completely different. Especially endocow was
disappointing. We splitted into teams, one trying new functions, one (ie. me)
trying to find the offset into DNA, where the coordinates of that object was
stored, and one group repositioning the objects to the right spot and
submitting to server.
Due to the evil mutations, encryption and a virus in the DNA, we did not get cloud, sun, caravan, hills, etc. working. We simply had not much time left and it was a time-consuming task to grep the logs for calls of methods and then skip backwards to find the last call of moveTo and then locate it's parameters. Our prefix steadily improved on Monday morning. But we were getting more and more tired. Even the triple espresso that Markus brew couldn't keep me awake for a long time. At the end I had the strange idea to rotate the wind mill by patching the sine and cosine table, which even worked! I had to put the patch into an unused function though, to prevent the sky background from moving, though.
Result
Here is the final result that we submitted:

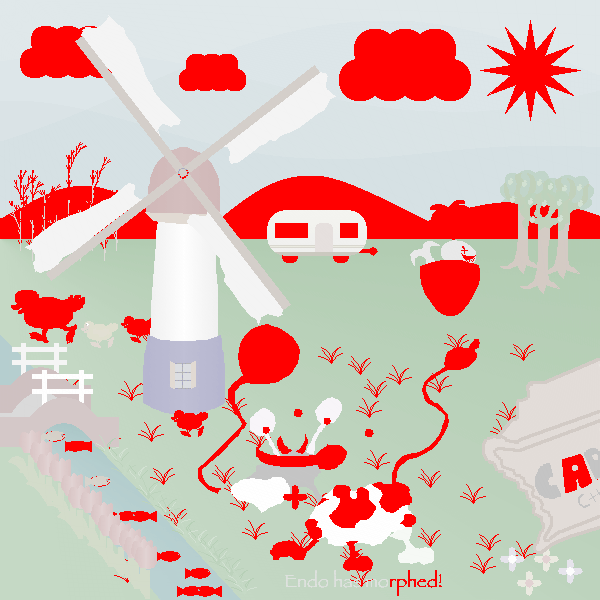
At the end we scored 9th with a risk of 543163 and a survival chance of 38.4472% with this prefix of length 4583.
When we finished the contest, I thought we have taken a sneaky approach by changing parts of the code, instead of setting the right variables and solving the riddles from the manual pages, e.g. searching for the steganography. We actually tried manipulating the variables but mostly with few or no success. I felt directly modifying the functions was a bit like cheating, however, it helped much improving our score. But reading the postmortems of other top fifteen teams, it turned out, that they did the same. Only later I found out, that this was the expected approach. For example, there is no other simpler way to recolor the cargo box, than by hacking the RNA commands as we did. The hints hidden in the manual pages are needed to reconstruct broken or encrypted methods like the clouds, the sun, the hills, the caravan or find the correct seed to grow the weed or the grass.
The lessons from the contest are the following. Read the specification completely! I found the remark about linear time algorithm only, when the first approach was too slow. Also I should have concentrated earlier on the part where I am good at: Reverse Engeneering. I could have started with the tools generating call graphs, automatically annotating instructions etc. and maybe even a disassembler on the second day, instead of writing a C program that would never be used. Having a second reference implementation is handy for bug tracking, but the first slow one was sufficient for this purpose. I should have trusted the rest of the team that they would finish their implementation in time. At the end the team work was better and everyone had a separate task. In hind-sight, it is also very bad that we ignored the prefix that appeared in our selftest. This can happen, even with a team of six people.
For those who want to know the ‘right’ solution, I have a short prefix reproducing the image completely. The organizers of the content did not produce such a prefix themselves. My guess is that the target image was drawn by a modified version of Endo's DNA, or even from their Haskell sources that they automatically translated to DNA.